-
Notifications
You must be signed in to change notification settings - Fork 3
Home
Welcome to the DAISIErobustness wiki. Here you will find a description of the DAISIErobustness project, technical details related to its implementation as an R package and instructions on how to use the package and replicate studies that use it (currently under review).
A short description of DAISIErobustness, its pipeline, and the broader DAISIE model can be found on this page. Follow the links on the sidebar or below to reach the sections on details about:
- Using and setting up the
Rpackage - Technical details of the package implementation
- Using the package on a HPCC (such as the University of Groningen Peregrine HPCC)
DAISIErobustness is an R package for testing of the robustness of the island biogeography model "DAISIE" (Dynamical Assembly of Island biota by Speciation, Immigration and Extinction) to more complex and potentially more realistic evolutionary models. Different measures of error of number of species, endemics, non-endemics and evolutionary trajectories are used to determine whether the alternative models can influence the inference capabilities of the current DAISIE model.
This is achieved via a pipeline which comprises the main functionality of the DAISIERobustness R package, containing all the necessary code to run it.
The DAISIErobustness project and package have been used to study the robustness of the DAISIE model to deviations from standard assumptions due to the influnece of geodynamics and traits on island biodiversity.
DAISIE is an evolutionary island biogeography model that allows for the estimation of diversification rates and other relevant diversification parameters on islands using phylogenetic data. These are:
- Cladogenesis rate
- Anagenesis rate
- (Clade-specific or island-wide) carrying capacity
- Migration rate
- Extinction rate
The estimation of such parameters is achieved using Maximum-Likelihood optimization, following the DAISIE likelihood functions described in published literature1. The performance of DAISIE inference given the amount and quality of data has been studied as well. It was shown to perform well, particularly when phylogenetic data is available2.
Furthermore, DAISIE includes simulation code allowing for the simulation of data following the above mentioned parameters.
DAISIErobustness main feature consists of a pipeline designed to measure the error one creates when extending the standard DAISIE model with new features. Examples of such new additions include the modelling of island ontogeny, as per the General Dynamic Model3, sea level changes4, and continental scenarios. The error measure is obtained by simulating and comparing DAISIE data using simulation code that builds upon the existing DAISIE simulations by including geodynamic processes.
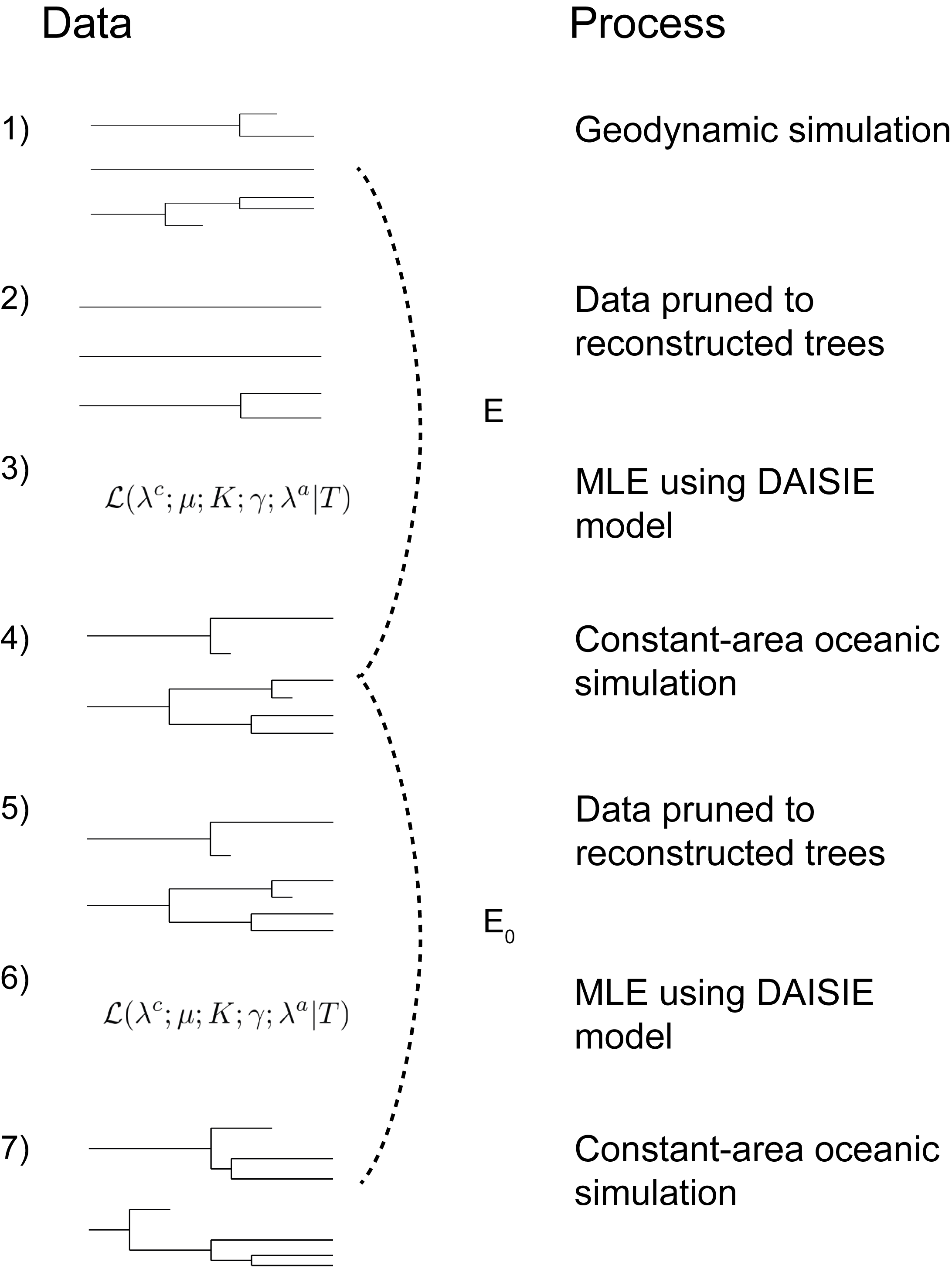 Schematic representation of the robustness pipeline.
Schematic representation of the robustness pipeline.
- Island phylogenetic data is produced by a geodynamic simulation.
- Reconstructed phylogenetic data is produced by pruning (removing) all extinct lineages.
- DAISIE maximum likelihood estimation (MLE) of parameters, which assumes no geodynamics, is applied to data from step 2.
- A constant-area oceanic simulation (i.e. the original DAISIE method) is run with the parameters estimates from step 3 as generating parameters.
- Phylogenies are pruned to reconstructed trees.
- The DAISIE MLE is applied to data from step 5.
- A constant-area oceanic simulation is run with parameter estimates from step 6 as generating parameters. The inference error made on data from a geodynamic simulation (E, step 3) is compared with the baseline error (E0, step 6). The error E is calculated by comparing the geodynamic simulations with the first set of constant-area oceanic simulations for five metrics (dashed line). The baseline error E0 is obtained by comparing two oceanic simulations using the same five metrics (dashed line). The error made when inferring from geodynamic data but assuming the constant-area oceanic model in inference is the proportion of errors (E) that exceed the 95th percentile of the baseline errors (E0).
1Santos Neves, Pedro, Joshua W. Lambert, Luis Valente, and Rampal S. Etienne. "The robustness of a simple dynamic model of island biodiversity to geological and eustatic change" bioRxiv 2021.07.26.453064 (2021)
2A simple island biodiversity model is robust to trait dependence in diversification and colonization rates Xie, Shu, Luis Valente, Rampal S. Etienne bioRxiv 2022.01.01.474685 (2022)
3Valente, Luis M., Albert B. Phillimore, and Rampal S. Etienne. "Equilibrium and non‐equilibrium dynamics simultaneously operate in the Galápagos islands." Ecology letters 18.8 (2015): 844-852.
4Valente, Luis, Albert B. Phillimore, and Rampal S. Etienne. "Using molecular phylogenies in island biogeography: it’s about time." Ecography 182 (2018): 820.
5Whittaker, Robert J., Kostas A. Triantis, and Richard J. Ladle. "A general dynamic theory of oceanic island biogeography." Journal of Biogeography 35.6 (2008): 977-994.
6Fernández‐Palacios, José María, et al. "Towards a glacial‐sensitive model of island biogeography." Global Ecology and Biogeography 25.7 (2016): 817-830.